Introduction
Fits a curve of "competition" of binding by an allosteric modulator, based on the ternary complex model. Note that this model assumes the allosteric modulator is present in excess, so is not depleted by binding to the receptors. Since it binds to a different site than the radioligand, the term 'competition' is not apt, but we list it here because the experimental design is the same as used for competitive binding.
Step by step
Create an XY data table. Enter the logarithm of the concentration of the unlabeled modulator (in nM) into X, and specific binding into Y in any convenient units.
From the data table, click Analyze, choose nonlinear regression, choose the panel of Competition Binding equations, and choose Allosteric modulator titration.
You must constrain two parameters to constant values based on your experimental design:
•RadioligandNM is the concentration of labeled ligand in nM. A single concentration of radioligand is used for the entire experiment.
•HotKdNM is the equilibrium dissociation constant of the labeled ligand in nM.
Also consider constraining Y0 (radioligand binding in the absence of modulator) to a constant value.
Model
AlloNM=10^(X+9)
KbNM=10^(logKb +9)
alpha=10^logAlpha
KAppNM=HotKDnm*(((1+(AlloNM/KBNM))/(1+alpha*(AlloNM/KBNM))))
HotOccupancy = RadioligandNM/(RadioligandNM + HotKDnm)
Y=(Y0/HotOccupancy)*(RadioligandNM/(RadioligandNM + KAppNM))
Interpret the parameters
Kb is the equilibrium dissociation constant (Molar) of modulator binding.
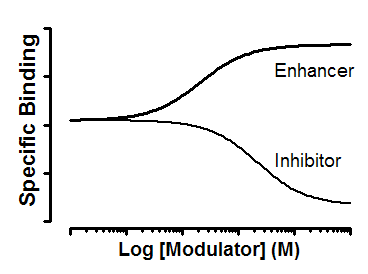
Alpha is the ternary complex constant. When alpha=1.0, the modulator does not alter binding. If alpha is less than 1.0, then the modulator reduces ligand binding. If alpha is greater than 1.0, then the modulator increases binding.
Y0 is the radioligand binding in the absence of modulator. Consider constraining this to a constant value.
Notes
This model is designed to analyze data when the unlabeled compound works via an allosteric site. Since the labeled and unlabeled ligands are acting via different sites, it is inappropriate (and incorrect) to refer to these types of experiments as “competition binding assays”. In some cases, in fact, the allosteric modulator enhances radioligand binding.
The model is written to fit the logarithm of alpha, rather than alpha itself. This is because alpha is asymmetrically (all values from 0 to 1 mean that the modulator decreases binding, while all values from 1 to infinity mean that the modulator enhances binding. On a log scale, its values are more symmetrical, so the confidence interval computed on a log scale (as Prism does) are more accurate.
The Y axis plots specific binding. Even at very high concentrations of inhibitor, the specific binding does not descend to zero. This is the nature of allosteric inhibition. If alpha is very high, then the binding is inhibited almost to zero. If alpha is not so high, then the maximum inhibition is more modest. For example, if alpha=3, the maximum inhibition is down to 33%.
This model assumes that the allosteric modulator is present in excess, so the concentration you added is very close to its free concentration. This model does not apply when the concentration of allosteric modulator is limiting (as it is when G proteins alter agonist binding to many receptors). No explicit model can handle this situation. You need to define the model with an implicit equation (Y on both sides of the equals sign) and Prism cannot handle such equations.
Reference
A. Christopoulos and T. Kenakin, Pharmacol Rev, 54: 323-374, 2002