How can I determine the Kcat of an enzyme?
Computing Kcat by hand
If you plot enzyme velocity as a function of subtrate concentration, you can fit the data to the Michaelis-Menten equation to determine the Km and Vmax.
The Vmax is the maximum enzyme velocity extrapolated out to very high concentrations of substrate. It is expressed in the same units you used to enter your Y values (enzyme activity). Usually it is straightforward to express this (or convert to ) moles/minute/mg of protein.
If you know the concentration of enzyme sites you've added to the assay (Et) then you can calculate the catalytic constant Kcat. It is defined to equal Vmax/Et.
Vmax and the Y values (enzyme velocities) are expressed in units of concentration per time, and Et must be entered in those same concentration units. Distinguish the concentration of enzyme molecules from the concentration of enzyme sites. For example, if the enzyme is a dimer with two active sites, the molar concentration of sites is twice the molar concentration of enzyme.
When calculating Kcat, the concentration units cancel out, so Kcat is expressed in units of inverse time. It is the turnover number -- the number of substrate molecule each enzyme site converts to product per unit time.
Fitting Kcat with Prism
You can also determine the Kcat directly by fittng this model to your data. It is built in to Prism (starting with Prism 5) in the enzyme kinetics group of equations.
Y=Kcat*Et*X/(Km + X)
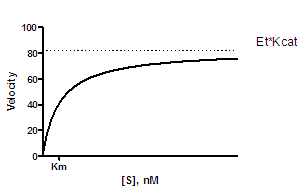
Y is enzyme activity, usually expressed as moles/minute/mg of protein.
Et is enzyme concentration. The Y values are entered in units of concentration per time, and Et must be entered in those same concentration units. It is impossible to fit both Kcat and Et, so you need to constrain Et to a constant value when fitting this equation.
Kcat is turnover number, expresed as number of substrate molecules turned into product per enzyme site per minute.
Km and X are both in the same units of substrate concentration.
Prism will fit Kcat and Km, each with a confidence interval.
Fitting the sample data
- From the Welcome dialog, choose XY and choose to use the Michaelis Menton sample data.
- Choose nonlinear regression and choose the kcat equation.
- On the constrain tab, constrain Et to equal 100.
- Prism reports that Km= 5.886 with a 95% confidence interval ranging from 3.933 to 7.839. The best fit value of kcat is 13.53 with a 95% confidence interval ranging from 11.97 to 15.09.
Keywords: enzyme kinetics















