Example
When an experiment has two possible outcomes, the results are expressed as a proportion. Since your data are derived from random sampling, the true proportion in the overall population is almost certainly different than the proportion you observed. A 95% confidence interval quantifies the uncertainty.
For example, you look in a microscope at cells stained so that live cells are white and dead cells are blue. Out of 85 cells you looked at, 6 were dead. The fraction of dead cells is 6/85 = 0.0706.
The 95% confidence interval extends from 0.0263 to 0.1473. If you assume that the cells you observed were randomly picked from the cell suspension, and that you assessed viability properly with no ambiguity or error, then you can be 95% sure that the true proportion of dead cells in the suspension is somewhere between 2.63 and 14.73 percent.
How to compute the confidence interval with Prism
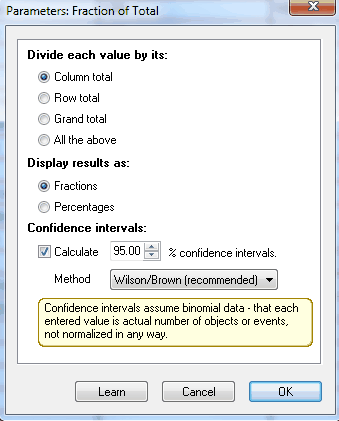
1.Create a new table formatted for parts of whole data.
2.Enter data only into the first two rows of column A. Enter the actual number of times each outcome occurred. For the example, enter 6 into the first row (number of blue dead cells) and 79 into the second row (number of white alive cells). Don't enter the total number of events or objects you examined. Prism will compute the total itself.
3.If you have more proportions that you wish to compute a confidence interval for, enter them into more columns of the data table.
4.Click Analyze, and choose the Fraction of Total analysis.
5.Choose to divide each value by its column total, and check the option to compute 95% confidence intervals. Choose whether you want to see the results as fractions of percentages.