Introduction
You can determine the equilibrium dissociation constant of an unlabelled ligand by measuring its competition for radioligand binding.
Step by step
Create an XY data table. Enter the logarithm of the Molar concentration of the unlabeled compound into X, and binding into Y. If you have several experimental conditions, place the first into column A, the second into column B, etc. Use subcolumns to enter replicates.
From the data table, click Analyze, choose nonlinear regression, choose the panel of Competition Binding equations, and choose Two sites - Fit Ki.
You must constrain three parameters to constant values based on your experimental design:
•HotNM is the concentration of labeled ligand in nM. A single concentration of radioligand is used for the entire experiment.
•HotKdNMHi is the equilibrium dissociation constant of the labeled ligand for the high-affinity site in nM.
•HotKdNMLo is the equilibrium dissociation constant of the labeled ligand for the low-affinity site in nM.
Model
logEC50Lo=log(10^logKiLo*(1+HotNM/HotKdNMLo))
logEC50Hi=log(10^logKiHi*(1+HotNM/HotKdNMHi))
Span=Top - Bottom
Part1=FractionHi*Span/(1+10^(X-LogEC50Hi) )
Part2=(1-FractionHi)*Span/(1+10^(X-LogEC50Lo) )
Y=Bottom + Part1 + Part2
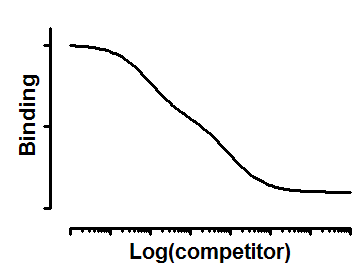
Interpret the parameters
Top and Bottom are plateaus in the units of Y axis.
FractionHi is the fraction of all the sites that have high affinity for the competitor.
logKi_Hi and logKi_Lo are the logarithms of the two molar Ki values.
Notes
This model fits the two log(Ki) values of the unlabelled ligand directly. It does not report the IC50s, so you do not need to apply the Cheng and Prusoff correction(1). Instead you enter the concentration of radioligand and its Kd as constants, and Prism directly fits the Ki of your cold compound. If you want to fit the two IC50 values instead of the Ki values, use a different equation.
The analysis assumes that you know the affinity of both sites for the labeled ligand. In many cases, the radioligand has the same affinity for both sites. In that case, simply enter that value twice. If the two sites have different affinities for the labeled ligand, enter both values (determined from other experiments). Watch out for the labels. The constant KdHi is the Kd of the hot ligand for the receptors with the high affinity for the unlabeled ligand, and KdLo is the Kd of the hot ligand for the receptors with lower affinity for the unlabeled ligand. So KdHi may be larger or smaller than KdLo.
This analysis assumes that the binding is reversible and at equilibrium. It also assumes that the labeled and unlabeled ligands compete for the same binding sites.
1. Cheng, Y. and Prusoff, W. H. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol, 22: 3099-3108, 1973.